Level 9: Multi-Agent "Games" to Solve X
Let AI Agents Drive Scientific Discovery
Welcome to The Gameful Scientist! This newsletter explores the intersection of scientific discovery and creativity. Enjoy!
A brief reminder on Project Grow:
We’re building a digital Los Alamos.
Inspired by Stanford and Google's Smallville, this is a multi-agent environment where AI agents - from scientists to biosecurity experts - collab on “Moonshot” projects. It demonstrates how multi-agent systems can generate emergent discoveries through diverse domain expertise and perspectives.
With an added layer of fun on top.
Let’s kick off this article with some level design.
Welcome to Viriditas: A digital town that serves as the core setting of Project Grow. It’s inspired by two things:
1. The Spirit of Los Alamos: Modeled after the secret town in New Mexico (but with more foliage) where the world’s greatest minds gathered to build the atomic bomb during WWII. Imagine this virtual town populated by the brightest and most capable digital beings, all focused on tackling “X”—the ambitious, crazy, and wonderful moonshot projects in biotech.
2. ’s Essay. This is a fantastic read! He writes about embracing biological growth to unlock profound abundance.
If AGI is a rocket ship with any chance of launching sentient life into the Cosmos, something deeply important and beautiful would be missing if it weren’t launching biological organisms.
Why a digital Los Alamos?
We live in a world of atoms, not bits. I understand that. But “doing science” through our current systems is too slow and constrained by the biophysical limitations of our machines and our experiments. I’d argue that to push scientific discovery at pace that will allow us to realize a solarpunk future in our life time, we need to lean more into the digital world. And with foundational models for bio coming online, it’s now possible to make real-world progress in a simulated environment. AI has completely reimagined how we can approach the life sciences. Project Grow is built with this thesis, that we can automate much of discovery through AI. And my hope is that we can bridge real-world science with storytelling, creating a space where we can dream AND actualize those dreams.
The Game-Like Approach
Project Grow leans heavily on game mechanics, not because it’s a “game” in the traditional sense, but because I believe using game mechanics is the best way to tackle these challenges.
Guiding Question: If superintelligence is accessible via an API, what would you build with it?
Multi-Agent Systems
I touched on multi-agent environments briefly in my last article, but let’s dig a little deeper.
At its core, Project Grow is a multi-agent environment, populated by AI agents that can think and act. These agents aren’t just chatbots; they make decisions autonomously, use tools, and perform tasks without direct human oversight.
Here’s why digital agents have unique advantages over humans:
No physical constraints
Rapid iteration
Perfect memory/knowledge sharing
24/7 operation
Creating these autonomous agents isn’t easy, but it’s achievable—and the safest way to deploy them is in a digital environment. Why? Because if they mess up, the failure is contained within a “soft-edge case.” In contrast, a medical AI agent analyzing patient data autonomously would be a “hard-edge case,” where mistakes can have severe real-world consequences. Like a patient dying.
Examples of Multi-Agent Systems: OpenAI’s hide-and-seek simulation, Stanford’s Smallville and Altera’s Project Sid in Minecraft show what’s possible.
Project Sid, in particular, is super exciting. It deployed over 1,000 agents to simulate a civilization. They demonstrated that agents can coordinate and perform tasks like mining and crafting, which are surprisingly translatable to real-world challenges.
But I want to design these environments, specifically for scientific discovery.
So here are some multi-agent projects for research that are quite incredible:
They’ve all come out within the last couple of months.
These projects are converging on a common idea: digital environments filled with collaborating AI agents can drive scientific discoveries.
This becomes an automated discovery system. And this is the space that I’ve been building in. For the AI people. I’m building “Level 4”.
Building Project Grow
In Project Grow, the most ambitious moonshot ideas live in digital “sandbox” environments, where our AI agents can go brainstorm together, and (hopefully) build amazing solutions.
One key question I kept running into was:
“Do the agents need to be walking around in 3D?”
The short answer: not necessarily. They can just as easily be abstract programs behind the scenes. But I’ve found a consumer-friendly approach means giving them some sort of embodiment. People connect better with characters—even if they’re just wearing lab coats and wandering around a stylized digital town. That’s what I’ve tried to do here. It definitely took me some heavy lifting in Blender and Unity (and I’m still not 100% happy with the aesthetics). If you’re a 3D environment or character artist, please ping me. Help is always welcome!
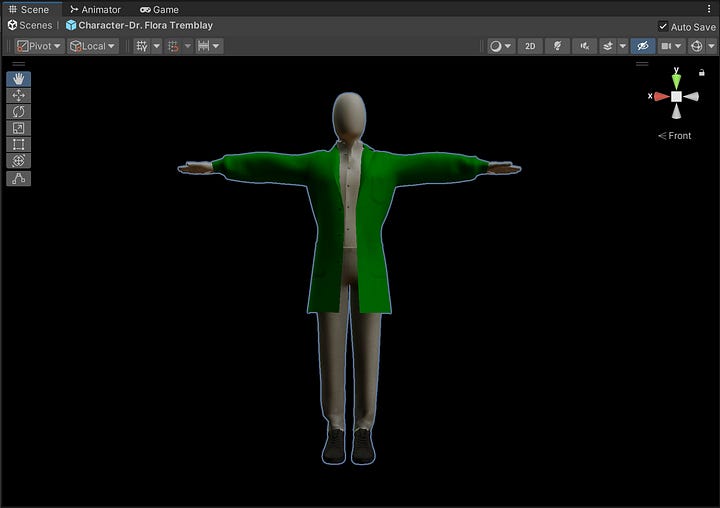

Meet the Agents
In the current prototype, it’s limited to 10 agents—though theoretically, we could scale up to thousands.
These are the roles:
• Scientist (non-playable): Fundamental research
• Biohacker: DIY biotech & distribution
• Economist: Viability analysis
• Creative: Artistic/creative biotech
• Doctor: Medical applications & safety
• Engineer (non-playable): Manufacturing & scaling
• Futurist: Outer space & off-world usage
• Biosecurity: Keeping watch for emerging threats
• Founder: Building business models
• Journalist (non-playable): Our real-time observer
Most are “autonomous” NPCs who can initiate or join collaborations. They share knowledge, propose wacky ideas, and react to each other’s breakthroughs. The Journalist agent logs these events into Google Sheets and compiles a “Whitepaper” in Google Docs, capturing the simulation’s evolving narrative.
Collab-Generated Breakthroughs
So how do we get big discoveries in the game?
Agents Collab: Let’s say our Scientist pairs with the Biohacker. They chat, bounce domain knowledge back and forth, and eventually produce a new “Breakthrough” idea—basically an unexplored angle or novel concept that’s relevant to the current moonshot.
Logged at the Biofoundry: We store this “Breakthrough” event in the in-game “Biofoundry” (the place where new ideas get turned into prototypes). You can think of it like an event-driven architecture: collaborations generate “events,” these events create “Breakthroughs,” and everything is logged in a central log.
Human-in-the-Loop: We deliberately added a step where you, the human, choose whether to “implement” a breakthrough. Partly because we’re rate‐limited by certain AI calls (like ESM3), but also because it’s more fun to let users direct the show. You can browse recent breakthroughs in the Biofoundry and decide which ones are worth building out.
Inspired by SciAgents: Our Approach to Breakthroughs
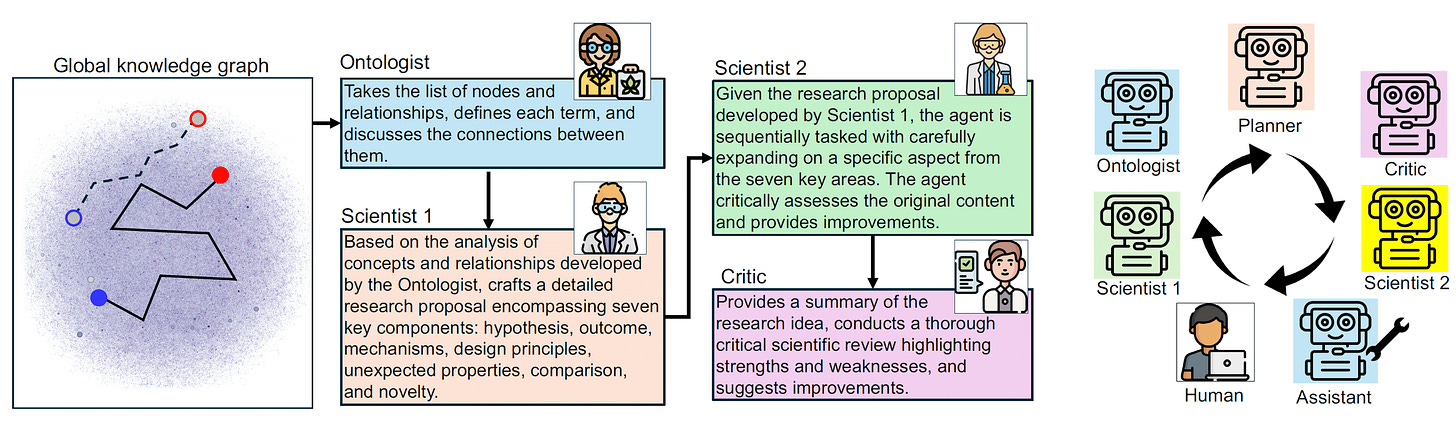
We drew inspiration from SciAgents, an agentic AI system developed by Markus J. Buehler’s lab at MIT. SciAgents uses large‐scale knowledge graphs, adversarial expert pairs, and interdisciplinary reasoning to drive scientific discovery. The idea of AI autonomously exploring novel domains and uncovering hidden patterns in complex data really resonated with us.
For Project Grow, we’ve adopted a similar philosophy but simplified development by integrating OpenAI’s O1 reasoning model. O1 serves as the “brain” behind our agents’ breakthroughs. It helps our system interpret the context of each moonshot, weigh the domain knowledge contributed by the agents, and propose new, untested angles.
The process:
Whenever agents collaborate, O1 (combined with the domain expertise of the participating agents) generates a Breakthrough. This is often a novel hypothesis or conceptual direction based on the moonshot’s challenge. For instance:
• The Doctor might highlight an overlooked antibiotic potential.
• The Futurist might connect a biotech discovery to a space application.
• The Biosecurity expert might flag an emerging risk worth tackling.
These interdisciplinary sparks are logged as Breakthroughs in the Biofoundry, ready for the player (you!) to implement.
RAG + ESM3: Making Breakthroughs “Real”
When you do choose to “implement” a Breakthrough at the Biofoundry, we run a Retrieval‐Augmented Generation (RAG) step plus a two‐pass ESM3 design. Here’s how that works:
Domain Discovery: We have a curated database of 500 Pfam families (pulled from InterPro)—things that might be relevant to climate, space, food, defense, etc. We also used GPT4o to make the functional annotation more readable.
Find a Match: Our system (again using O1 reasoning) interprets your new Breakthrough and picks one or more domains from that database.
Mask the Sequence: We form a partial protein sequence template, something like ____{motif}____, letting ESM3 fill in the underscores with valid amino acids.
Pass A (track = “sequence”): ESM3 sees these underscores (_) and unrolls a new protein sequence.
Pass B (track = “structure”): ESM3 uses that final sequence to generate a predicted structure and outputs a pTM (confidence) score.
Log: We store it all—final sequence, pTM, domain info—in the simulation so you (and the other agents!) can see how stable or promising this design might be.
Iterative Discovery: Agents React
If the pTM is too low, or if an agent (like the Biosecurity expert) flags it as risky, the team might refine it again. We can mask part of the newly generated sequence (like _ in the middle) and have ESM3 propose a variant. Each iteration goes into the Biofoundry log as a new “Eureka.” Agents keep responding socially to these “Eureka” events based on their role. That interplay is what I love—the simulation is driven by the loop of:
Collaborations → Breakthroughs
Implementation → ESM3 + confidence
Agent Reactions → Possibly more re-collaboration or domain changes
And on it goes, building out a chain of designs—some dead ends, some brand‐new wonders.
Why This Matters
Sure, it’s still an early prototype. But we did recently get beta access to ESM3, so now our playful “Eurekas” are no longer purely imaginary. They’re actual, model‐generated protein sequences with predicted structures. That means we’re inching closer to actual scientific utility. For deeper details on how ESM3 works, check out
. He has a great ongoing series called “Understanding Protein Language Models”.An Example
Let’s look at how Project Grow tackles an ambitious challenge—bringing back the woolly mammoth. We have other moonshots in Climate, Space, Health, Fashion, Food and Defense. But let’s use the theme of de-extinction as an example:
Step 1: Collaboration Sparks a Breakthrough
Scientist Agent identifies gaps in mammoth hemoglobin DNA and collaborates with the Biohacker Agent to brainstorm solutions.
Together, they propose a breakthrough:
“Design mammoth hemoglobin optimized for extreme cold.”
The breakthrough is logged at the Biofoundry and awaits your (the player’s) decision to implement it.
Step 2: Implementing the Breakthrough
You decide to implement the breakthrough. Here’s what happens next:
OpenAI’s O1 reasoning model analyzes the breakthrough and identifies a relevant Pfam domain, such as the Globin domain.
A masked sequence template is generated: ____[globin motif]____
Step 3: Protein Design with ESM3
• ESM3 runs in two passes:
1. Pass A (Sequence Track): Fills the masked sequence to generate a plausible mammoth hemoglobin protein.
2. Pass B (Structure Track): Predicts the structure and outputs a pTM confidence score (e.g., 0.78).
The new protein design is logged as a Eureka, complete with its sequence and confidence score.
Step 4: Agent Reactions and Emergent Creativity
The Eureka Moment ripples through the simulation, prompting unexpected reactions from the agents. Here’s where things get interesting:
Biohacker Agent:
Suggests testing the protein’s thermal stability, hypothesizing it could be adapted for other cold environments, such as cryopreservation technologies or food storage.
Creative Agent:
Proposes using the protein’s unique structural properties in fashion:
“What if we integrate this mammoth-inspired protein into a biodegradable fabric? Imagine winter gear that adapts to extreme cold like mammoth fur!”
Logs a secondary breakthrough idea:
“Engineered protein fibers for extreme-weather textiles.”
Futurist Agent:
Explores the possibility of using the protein in terraforming experiments for Mars, hypothesizing that a modified version could enhance cold-resistance in plants or other organisms in extreme environments.
Biosecurity Agent:
Flags potential risks of introducing this protein into non-target species if used in the wild, prompting the team to pivot to testing isolated applications instead.
Step 5: Iterative Discovery and Cross-Domain Creativity
The initial hemoglobin protein, designed for mammoths, spins off into multiple new directions:
Fashion Innovation: The Creative Agent leads an effort to explore protein-based textiles. ESM3 is used again to modify the sequence for enhanced elasticity and durability while retaining thermal properties.
Cryopreservation: The Biohacker Agent repurposes the protein for food preservation, testing how it behaves in ultra-cold storage environments.
Mars Terraforming: The Futurist Agent initiates a new collaboration to adapt the protein for plants, creating a cold-tolerant symbiotic system for Martian agriculture.
Emergent Dynamics: Creative Outcomes in Project Grow
What makes this scenario exciting is the unexpected creativity that emerges. A protein initially designed for de-extinction becomes a launchpad for entirely new applications:
Fashion and Sustainability: Engineered proteins for adaptive, biodegradable winter gear.
Cryopreservation: Leveraging mammoth-inspired hemoglobin for low-cost food storage solutions in underserved regions.
Mars Applications: Using the protein to enhance cold tolerance in plants for off-world agriculture.
These emergent ideas highlight how the simulation fosters cross-domain thinking, turning even small discoveries into multi-disciplinary opportunities.
With emergent creativity baked into the system, Project Grow isn’t just about solving one problem—it’s about watching new possibilities unfold. The mammoth hemoglobin protein, originally designed for de-extinction, becomes a gateway to breakthroughs in sustainability, food storage, and space exploration. This is the power of multi-agent collaboration and AI-driven discovery: ideas that evolve beyond their original scope, revealing potential you’d never predict on your own. Eventually, this goes beyond a digital world. Print the sequence and build the thing in the physical world. In the world of atoms.
Disclaimer: A complete mammoth revival requires precise biological data that AI models alone can’t fully supply—such as high-quality DNA sequences, epigenetic markers, and insights into gene-environment interactions.
I’m trying to build Project Grow into something more than just a social simulation. This next generation is growing up in digital environments, and I believe platforms like this can be real research tools for anyone—not just scientists. Whether it’s designing mammoth habitats or lab-grown meat, the goal is simple: simulate, strategize, and create real-world impact.
Simulated worlds let us explore multiple futures for the technologies we build.
Most of you don’t know that I have a twin. He recently mentioned that I should stop calling this platform a “game”. Doesn’t resonate with people. Instead call this a simulation. Agreed.
Thanks for reading The Gameful Scientist!
Feel free to contact me here or chat with me on Twitter @ATrotmanGrant :)











This is a really creative idea! Love the use of agents to generate ideas for ESM3 to go validate.
I'm curious - when a Eureka moment stored in the Biofoundry and selected by the user for simulation, are you using the initial RAG to identify relevant gene ontology labels in addition to the domains? For example, building off the wooly mammoth use case, you could condition the protein generation using the gene ontology label GO:0009631, which describes "any process that increases freezing tolerance of an organism in response to low, nonfreezing temperatures".